Sequence Determination
Sequence Determination Assignment Help | Sequence Determination Homework Help
Sequence determination
There are several methods for the determination of primary structure proteins . The most elegant method was developed by sanger and his associates in 1955, who elucidated the complete sequence of amino acids in the polypeptide hormone, insulin. This work was rewarded with the nobel prize in1957.
Amino acid composition In order to determine types of amino acids and their molar ratio in a polypeptide chain, complete hydrolysis may be carried out by heating a protein with 6N HCI at 110o in an evacuated sealed tube for 6 to 48 hours. The resulting mixture, from which excess of HCI is removed, is called protein hydeolyzate containing a mixture of all constituent amino acids. Acid treatment cleaves peptide bonds to free the modified, e.g., tryptophan is destroyed while glutamine and asparagine are modified to their respective acids. However, amino acids thus obtained may be quantitatively determined by column chromatography or amino acid analyzer. The total amino acid content of β-lactogloulin (a milk protein) was determined in this manner. Proteins can be hydrolyzed by boiling with alkali, but this method is unsuitable since it causes racemization of most and destruction of some amino acids. The amino acids that are destroyed are cysteine,serine, threonine and cystine.
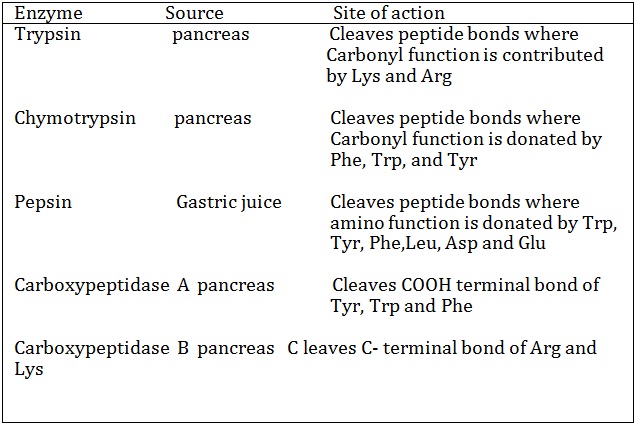
cleaving sites of some proteolytic enzymes used in partial hydrolysis
cleaving sites of some proteolytic enzymes used in partial hydrolysis
Determination of N-terminal Amino acid
1. Sanger,s method The N-terminal amino acid is identified by the reaction with 1-fluoro-2, 4-dinitrobenzene (FDNB) yielding dinitrophenyl residue of the N-terminal amino acid. The DNP-peptide is then hydrolyzed by 6N HCI so that all peptide bonds are cleaved leaving the DNP-derivatives. Since this method involves hydrolysis of peptide, it cannot be used for stepwise degradation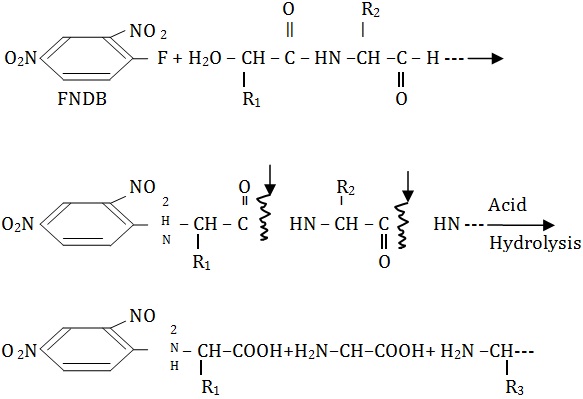
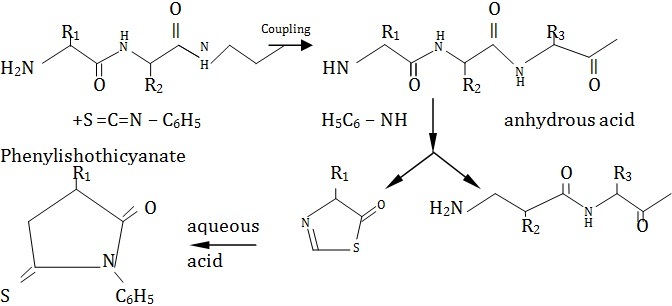
2. Edman degradation This method is very useful in determining the N-terminal amino acid. The peptide fragment is treated with phenylisothiocynate at pH 8 to yield phenylthicarbamyl derivative at the N-terminal.The derivative is treated with acid in organic solvent so that N-terminal amino acid undergoes cyclization to produce phenylthiohydantoin which is cleaved form the peptide fragment. Thiohydantoin derivative can be identified by paper chromatography and the peptide is further subjected to the same treatment, each time forming a thiohydantoin derivative from the amino end.

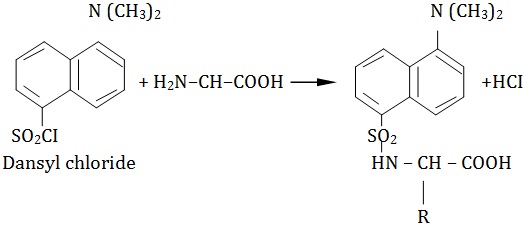
1. Dansyl chloride Dansyl chloride regent is used for determination of N-terminal residue in alkaline condition. The N-terminal residue forms a yellow fluorescent derivative which can be easily detected in minute amounts. The derivative is also resistant to acid hydrolysis of polypeptides.
For more help in sequence Determination please click the button below to submit your homework assignment