The Promoter Gene
The Promoter Gene Assignment Help | The Promoter Gene Homework Help
The Promoter gene
The promoter gene is continuous with the operator gene. This sequence extends from the termination codon (ACT on DNA = UGA on mRNA) of the structural gene, and includes the promoter and operator genes. The promoter region a sequence a two-fold symmetry in which sections of DNA on either site of a particular point are symmetrical. The sequences in these sections are ‘palindromic’. The nucleotides of one strand go in one direction, while an identical sequence of nucleotides goes in the opposite direction in the other strand. It has been suggested that these symmetrical regions of DNA may be recognized by proteins having symmetrically arranged subunits. The CRP site contains this section of two-fold symmetry. The CRP site binds a protein called CRP (cyclic AMP receptor protein). This protein is essential for the binding of the enzyme RNA polymerase to the promoter. The CRP is a dimmer with two subunits and molecular weights of 45,000. In E.coli the CRP combines with cyclic adenosine monophosphate (cAMP) to form a CRP-cAMP complex. This complex binds to the promoter. The CRP has a strong affinity for DNA, and this affinity increases considerably when the protein combines with cAMP. Binding of the CRP-cAMP complex to the promoter enhances RNA polymerase attachment to the promoter and thus increases transcription and protein synthesis. This is known as positive control.It has been found that in E.coli and certain phages the RNA polymerase site and forms a stable complex, if such DNA molecules are treated with pancreatic DNase (an exzymes which breaks down DNA), ihe region of DNA forming the complex with RNA is protected from the action of DNase. This protected region is found to consist of 40 – 50 base pairs, and is not cleaved into smaller pieces by DNase.
The RNA polymerase site has a small region of high A.T pairs, with high G.C regions on either site.
These are (i) a recognition sequence (ii) a binding sequence and (iii) an mRNA initiation site.
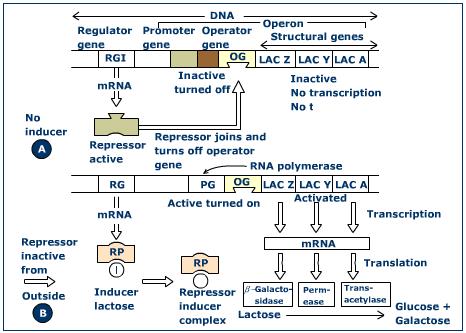
The recognition sequence is outside the polymerase binding site, i.e. the region of DNA protected against the action of DNase. According to the model, RNA polymerase firsts binds to DNA by forming a complex with the recognition sequence. It then binds with the binding sequence to produce the pre-initiation complex.
The binding site consists of a sequence of seven bases. These are present in a constant location in the protected fragments. The sequence of this set of seven bases has been found to be almost constant in the bacteria and phages studied. It never differs by more than two bases from the following sequences: 5’TATPuATG.
RNA initiation site. The start of mRNA transcription takes place on one of the two bases near the binding sequence. This starting site is including in the DNA fragment protected from DNase digestion. In the lac operon there is overlapping of promoter and operator sites. At least a part of the operator sequences appear in the polymerase binding site sequence.
In the promoter region, 20 base pairs to the left of the starting site of transcription, is a sequence 5’CCGG. This is the substrate for an endnuclease enzyme isolated from Hemophilus parainfulenzae, and is know as the Hpa II site. This site has been used as a reference point in studies of repressor and polymerase binding.
For more help in The Promoter gene please click the button below to submit your homework assignment.